BioFlow-Insight
Welcome to BioFlow-Insight, the tool that allows you to gain insight into your Nextflow workflow!
From your workflow’s source code, BioFlow-Insight generates graph structures that represent the different steps of your workflow, easily and without any configuration or the need to execute it. BioFlow-Insight analyses the code of your Nextflow workflow and then automatically reconstructs its structure. It is even capable of detecting different types of errors in the code. For more information on BioFlow-Insight’s functionalities, error handling, and guidelines, please refer to the Specification List.
Example of two different types of graphs generated by BioFlow-Insight:
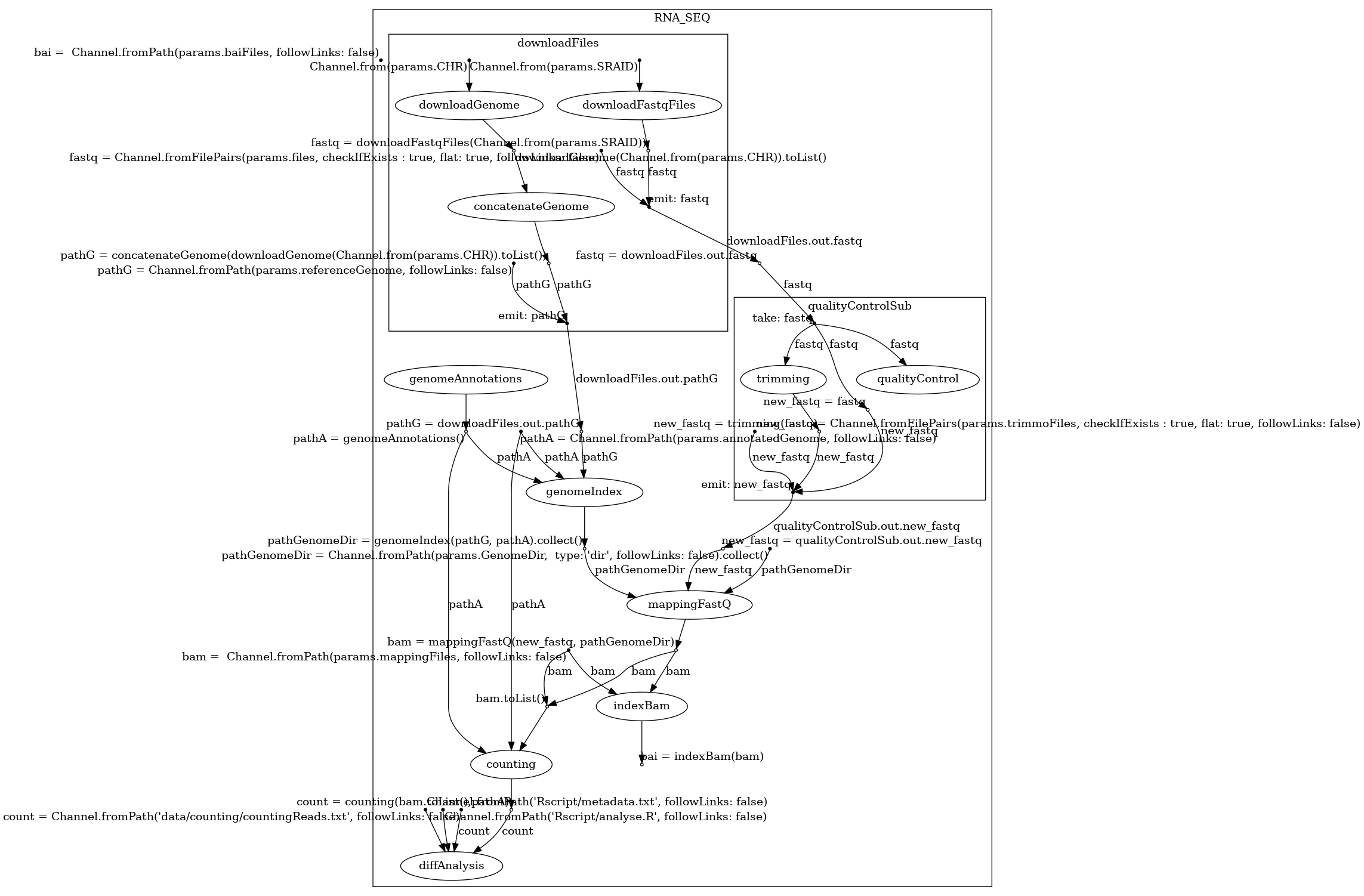
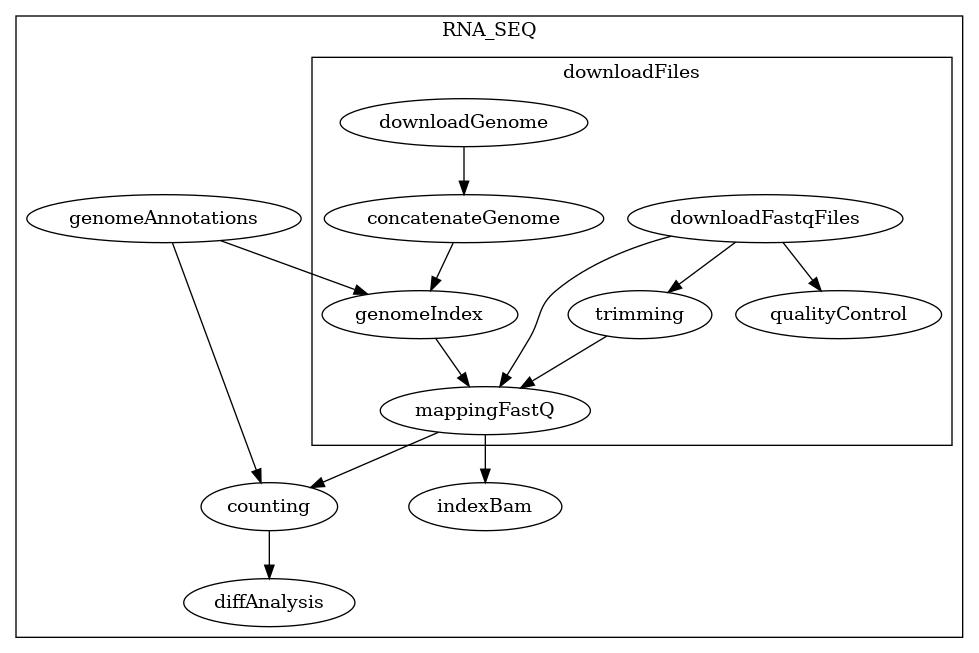
The specification graph on the left and the process dependency graph on the right. Both of these graphs provide a representation of the workflow https://github.com/George-Marchment/hackathon.
To cite BioFlow-Insight, please use the following publication:
George Marchment, Bryan Brancotte, Marie Schmit, Frédéric Lemoine, Sarah Cohen-Boulakia, BioFlow-Insight: facilitating reuse of Nextflow workflows with structure reconstruction and visualization, NAR Genomics and Bioinformatics, Volume 6, Issue 3, September 2024, lqae092, https://doi.org/10.1093/nargab/lqae092
BioFlow-Insight is an open-source tool under the GNU Affero General Public License, its source code can be found here: https://gitlab.liris.cnrs.fr/sharefair/bioflow-insight.
To test BioFlow-Insight functionalities, you can submit your workflows either by using a zip archive or the url of a git repository. Alternatively, you can try using the provided example workflows with the 'Submit from a git repository' functionality: